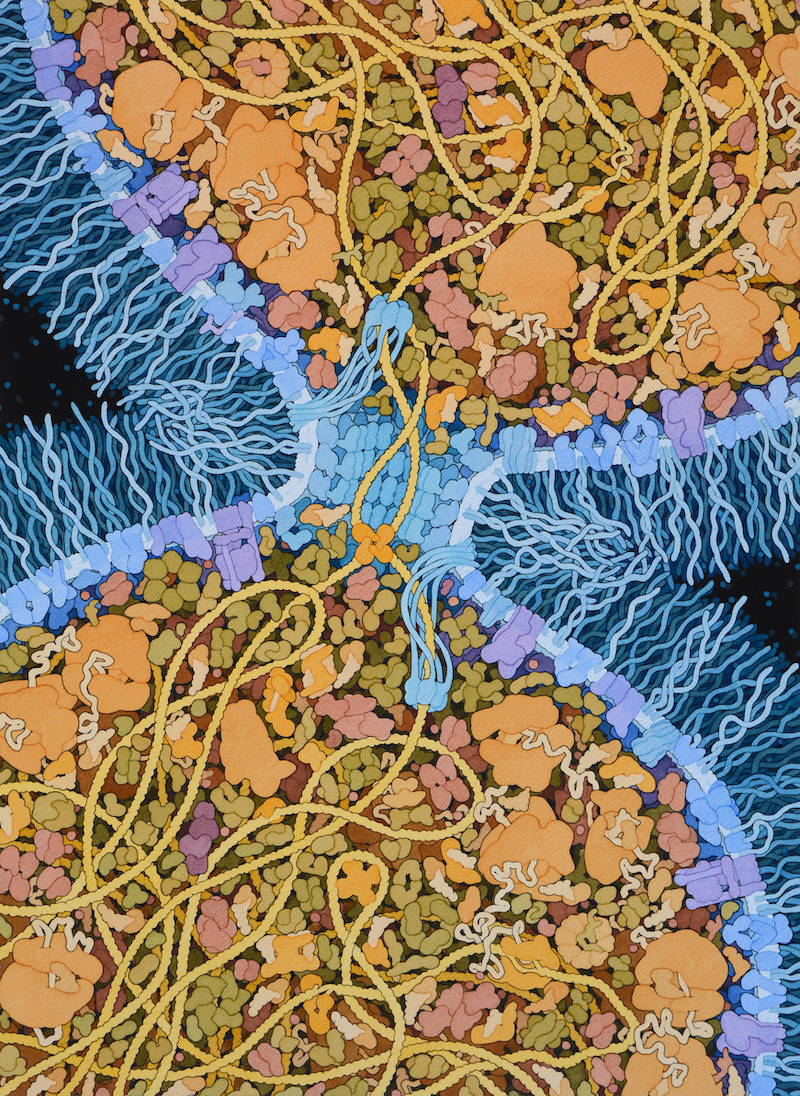
MOLECULAR LANDSCAPES BY DAVID S. GOODSELL
Last Universal Common Ancestor
Acknowledgement: Illustration by David S. Goodsell, RCSB Protein Data Bank. doi: 10.2210/rcsb_pdb/goodsell-gallery-035
This speculative painting depicts the primordial cell that gave rise to all life on Earth. The term “Last Universal Common Ancestor”, or LUCA, is based on the idea that this cell was so successful that it out-competed all of its rivals and ultimately was the parent for all cells that followed.
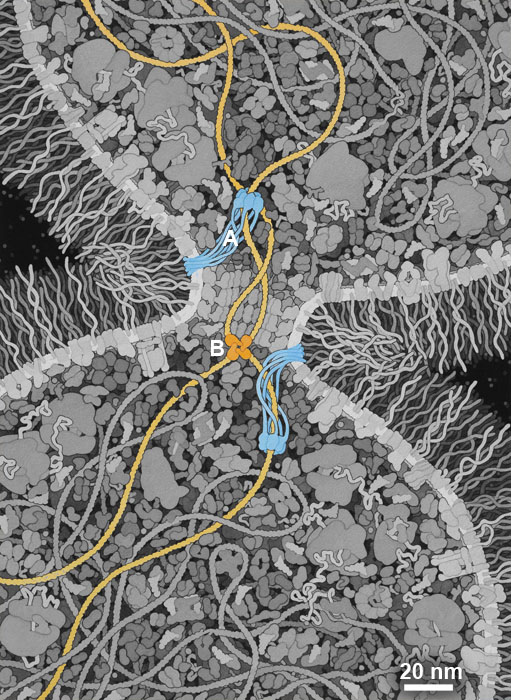
There are many theories about when and where LUCA arose, and what it looked like. I have chosen to highlight a property of LUCA that may have been a turning point in the evolution of a highly-competitive cell: the ability to efficiently separate two daughter DNA strands during cell division. I have included a simple mechanism used in modern bacterial cells that combines two proteins: (A) A DNA translocase (such as FtsZ) is attached to the septum between proteins that pulls DNA through its central pore until it finds (B) a recombinase protein (such as Xer recombinases) that recognize a specific site in the DNA and will ultimately separate the two daughter DNA strands. I have also chosen to give this hypothetical LUCA a simple cell wall composed of glycolipids to provide an additional competitive advantage.
The proteins included in this illustration are largely based on a recent analysis of modern bacterial genomes, looking for commonalities to give clues about the composition of LUCA (reference 3 below). This study sees LUCA as not using oxygen and able to synthesize all of its component biomolecules from raw materials available in the environment. It also sees LUCA living in hydrothermal vents and being dependent on conditions and molecules that are present there.
The painting was researched and rendered at a 2018 session of the Djerassi Resident Artists Program, kindly underwritten by Anne and Rodney Pearlman.
Selected References
1. Cornish-Bowden, A., and Cárdenas, M. L. (2017) Life before LUCA. J. Theor. Biol. 434, 68–74
2. Dubarry, N., and Barre, F.-X. (2010) Fully efficient chromosome dimer resolution in Escherichia coli cells lacking the integral membrane domain of FtsK. EMBO J. 29, 597–605
3. Weiss, M. C., Sousa, F. L., Mrnjavac, N., Neukirchen, S., Roettger, M., Nelson-Sathi, S., and Martin, W. F. (2016) The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 1, 16116
4. Williams, T. A., Szöllősi, G. J., Spang, A., Foster, P. G., Heaps, S. E., Boussau, B., Ettema, T. J. G., and Embley, T. M. (2017) Integrative modeling of gene and genome evolution roots the archaeal tree of life. Proc. Natl. Acad. Sci. 114, E4602–E4611
Learn more about other molecular landscapes:
Abiogenesis
This speculative painting depicts some of the processes that have been proposed for......

Cellulose Synthase
In this cross-section, a plant cell is shown at the bottom, with the plasma membrane......